41 maxquant label free quantification
PDF Label-free Quantification with PEAKS - Bioinformatics Solutions Inc. Label-free Quantification with PEAKS Wen Zhang, Ziaur Rahman, Yifang Chen, Lei Xin, and Baozhen Shan Bioinformatics Solutions Inc., Waterloo, Canada Aims Label-free quantification (LFQ) data analysis Summary: ... MaxQuant between the neighboring groups were shown in Figure 2. The UPS1 protein ratios Label-free data analysis using MaxQuant - Lazarus MaxQuant supports different "Quantitation Methods". The three main categories are label-free quantification, label-based quantification and reporter ion MS2 quantification. In this tutorial we have chosen label-free because we did not apply any specific labeling/quantitation strategy to the samples.
Accurate proteome-wide label-free quantification by delayed ... MaxLFQ is a generic label-free quantification technology that is readily applicable to many biological questions; it is compatible with standard statistical analysis workflows, and it has been validated in many and diverse biological projects. ... It is implemented in the freely available MaxQuant computational proteomics platform and works ...
Maxquant label free quantification
Label Free Quantitation using Maxquant LFQ (MaxLFQ) MaxLFQ is a generic label-free quantification technology that is readily applicable to many biological questions; it is compatible with standard statistical analysis workflows, and it has been validated in many and diverse biological projects. Our algorithms can handle very large experiments of 500+ samples in a manageable computing time. MaxQuant and MSstats for the analysis of label-free data In this training we will cover the full analysis workflow from label-free, data dependent acquisition (DDA) raw data to statistical results. We'll use two popular quantitative proteomics software: MaxQuant and MSstats. MaxQuant allows protein identification and quantification for many different kinds of proteomics data (Cox and Mann 2008). MQSS 2019 | L4: Label free quantification | Christoph Wichmann Quantitative proteomics long relied on stable isotope labels to compare the quantities of proteins across samples. Alternative label-free methods traditional...
Maxquant label free quantification. Comparative evaluation of label-free quantification strategies To compare the performance of widely used or recently developed label-free quantification strategies, we analyzed the data using seven distinct label-free quantification strategies ( Fig. 1 and Supplementary Table S1). MQ-SC and StPeter were based on spectral count, while other strategies were based on ion intensity. › articles › s41467/020/17033-7Rapid, deep and precise profiling of the plasma proteome with ... Jul 22, 2020 · We determined the median CVs for protein group quantification using MaxQuant (see Methods). The results ranged from 16.4 to 30.8% (Fig. 4b , Supplementary Table 4 , 5 ), which is in the range of ... Label-free data analysis using MaxQuant - GitHub Pages This stand-alone tutorial introduces the data analysis from raw data files to protein identification and quantification of two label-free human serum samples with the MaxQuant software. One sample is a pure serum sample, while the other sample has been depleted for several abundant blood proteins. One of the questions in this tutorial is to ... › articles › nprotThe MaxQuant computational platform for mass spectrometry ... Oct 27, 2016 · To enable LFQ go to the 'Label free quantification' page and select the 'LFQ' option. 28 Set the minimum ratio count that is required for a peptide feature to be used in the quantification of a ...
PDF Proteome-wide label-free quantification with MaxQuant - BSPR Label-free quantification Benchmark dataset •HeLa and E. coli cell lysates are mixed •Proteins were digested with trypsin. •In three replicates peptides were separated by isoelectric focusing in 24 fractions. •This was repeated with the same amount of HeLa, but E. coli lysate tripled. Comparative evaluation of label-free quantification strategies The selection of a data processing method for use in mass spectrometry-based label-free proteome quantification contributes significantly to its accuracy and precision. In this study, we comprehensively evaluated 7 commonly-used label-free quantification methods (MaxQuant-Spectrum count, MaxQuant-iB … MaxQuant MaxQuant MaxQuant is a quantitative proteomics software package designed for analyzing large mass-spectrometric data sets. It is specifically aimed at high-resolution MS data. Several labeling techniques as well as label-free quantification are supported. MaxQuant is freely available and can be downloaded from this site. Assessment of Label-Free Quantification in Discovery ... - PubMed Assessment of Label-Free Quantification in Discovery Proteomics and Impact of Technological Factors and Natural Variability of Protein Abundance J Proteome Res. 2017 Apr 7 ... Proteome Discoverer, Scaffold, MaxQuant, and Progenesis QIP, was benchmarked using their default parameters and some modified settings. Beyond this, the intersample ...
Accurate Proteome-wide Label-free Quantification by Delayed ... MaxLFQ is a generic label-free quantification technology that is readily applicable to many biological questions; it is compatible with standard statistical analysis workflows, and it has been validated in many and diverse biological projects. Our algorithms can handle very large experiments of 500+ samples in a manageable computing time. pharmazie.uni-greifswald.de › storages › uniMaxQuant Information and Tutorial - uni-greifswald absolute quantification: copy numbers or concentration of protein within a sample (challenge: different peptide species with various ionization efficiencies have to be quantitatively compared within the same sample) both: label-based (relative: e.g. SILAC, ICAT, TMT; absolute: e.g. AQUA) or label-free possible 1.4.2 label-free quantification Label-free data analysis using MaxQuant - Galaxy Training Network The three main categories are label-free quantification, label-based quantification and reporter ion MS2 quantification. In this tutorial we have chosen label-free because we did not apply any specific labeling/quantitation strategy to the samples. Figure 3: Overview of MaxQuant quantification methods MaxQuant is a quantitative proteomics software package designed for analyzing large mass-spectrometric data sets. It is specifically aimed at high-resolution MS data. Several labeling techniques as well as label-free quantification are supported.
› science › articleIntegrated Proteogenomic Characterization of HBV-Related ... Oct 03, 2019 · Introduction. Liver cancer ranks the fourth leading cause of cancer-related death worldwide (Villanueva, 2019).Hepatocellular carcinoma (HCC) accounts for about 85%–90% of all primary liver malignancies, and the largest attributable causes are chronic infection by hepatitis B virus (HBV) and hepatitis C virus (HCV) (Sartorius et al., 2015), along with alcohol abuse and metabolic syndrome.
Plant Phosphopeptide Identification and Label-Free Quantification by ... Phosphopeptide(s) identification and label-free quantification can determine dynamic changes of phosphorylation events in plants. Both MaxQuant and Proteome Discoverer are professional software tools used to identify and quantify large-scale MS-based phosphoproteomic data.
MaxQuant error at label free normalization step - Google Groups Then no issue with maxquant search. Good luck ... I am having a similar problem, but under my description it just says the file folder and "label free_normalization 0 Label free_normalization" and then it cuts off. I'm searching 180 files and it keeps happening. All files open fine in xcalibur, and most have searched fine in smaller searches.
adinasarapu.github.io › posts › 2018Quantitative proteomics: label-free quantitation of proteins Apr 04, 2020 · MaxQuant employs the MaxLFQ algorithm for label-free quantitation (LFQ). Quantification will be performed using razor and unique peptides, including those modified by acetylation (protein N-terminal), oxidation (Met) and deamidation (NQ). 2. Quality Control of MaxQuant Search (optional)
Comparative Evaluation of MaxQuant and Proteome Discoverer MS1 ... - PubMed MS1-based label-free quantification can compare precursor ion peaks across runs, allowing reproducible protein measurements. Among bioinformatic platforms enabling MS1-based quantification, MaxQuant (MQ) is one of the most used, while Proteome Discoverer (PD) has recently introduced the Minora tool. …
› dokumaxquant:start [MaxQuant documentation] Feb 26, 2019 · MaxQuant is a proteomics software package designed for analyzing large mass-spectrometric data sets. It is specifically aimed at high-resolution MS data. Several labeling techniques as well as label-free quantification are supported. MaxQuant is freely available and can be downloaded from this site.
Label-free data analysis using MaxQuant - Galaxy Training Network MaxQuant offers highly accurate functionalities for many different proteomics quantification strategies, e.g. label-free, SILAC, TMT. Blood is a commonly used biofluid for diagnostic procedures. The cell-free liquid blood portion is called plasma and after coagulation serum.
MQSS 2019 | L4: Label free quantification | Christoph Wichmann Quantitative proteomics long relied on stable isotope labels to compare the quantities of proteins across samples. Alternative label-free methods traditional...
MaxQuant and MSstats for the analysis of label-free data In this training we will cover the full analysis workflow from label-free, data dependent acquisition (DDA) raw data to statistical results. We'll use two popular quantitative proteomics software: MaxQuant and MSstats. MaxQuant allows protein identification and quantification for many different kinds of proteomics data (Cox and Mann 2008).
Label Free Quantitation using Maxquant LFQ (MaxLFQ) MaxLFQ is a generic label-free quantification technology that is readily applicable to many biological questions; it is compatible with standard statistical analysis workflows, and it has been validated in many and diverse biological projects. Our algorithms can handle very large experiments of 500+ samples in a manageable computing time.
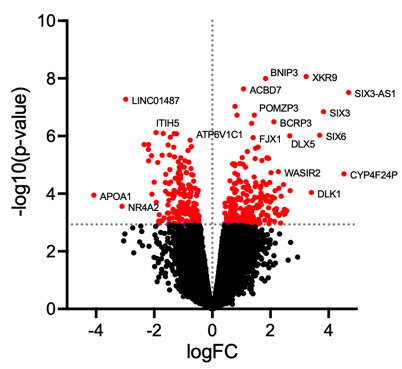






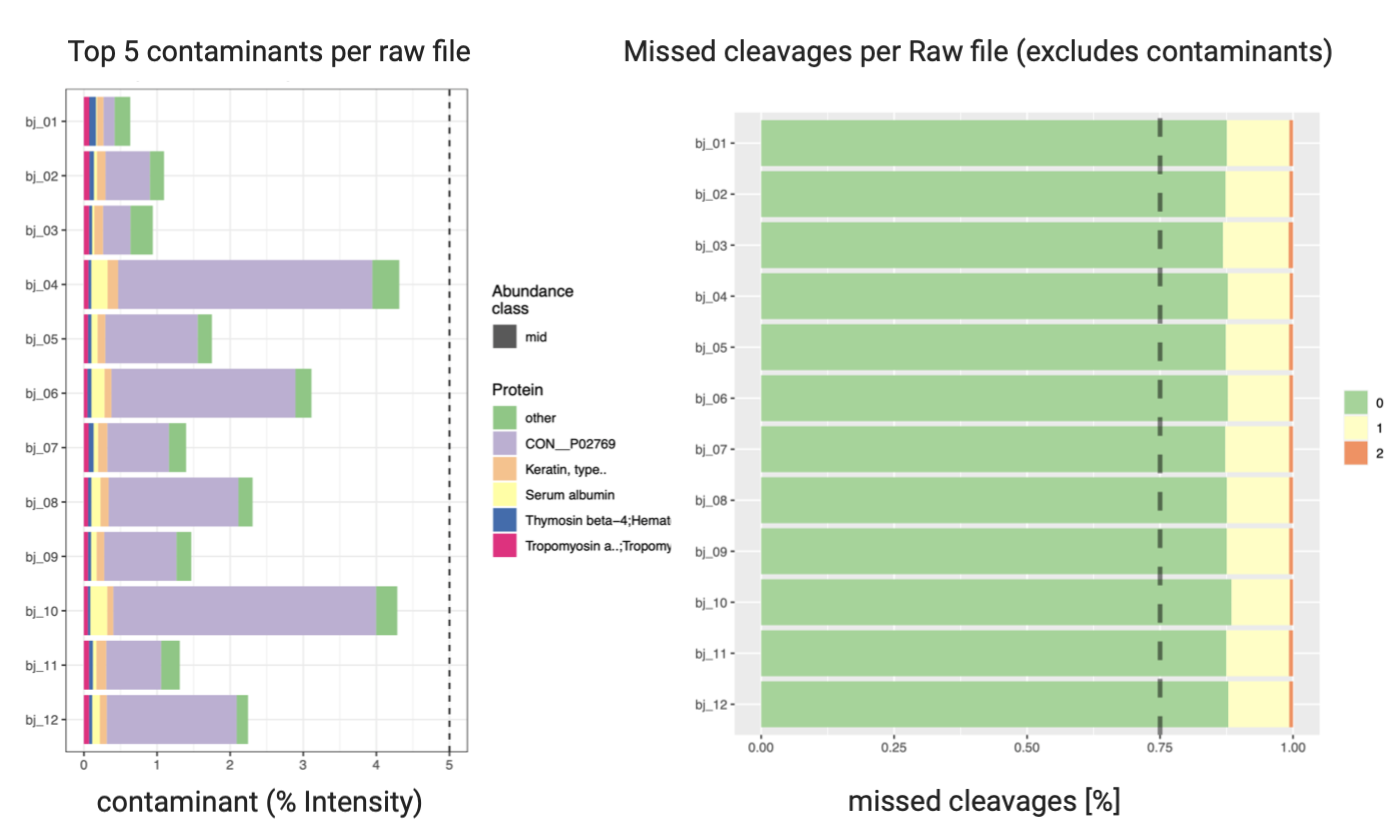



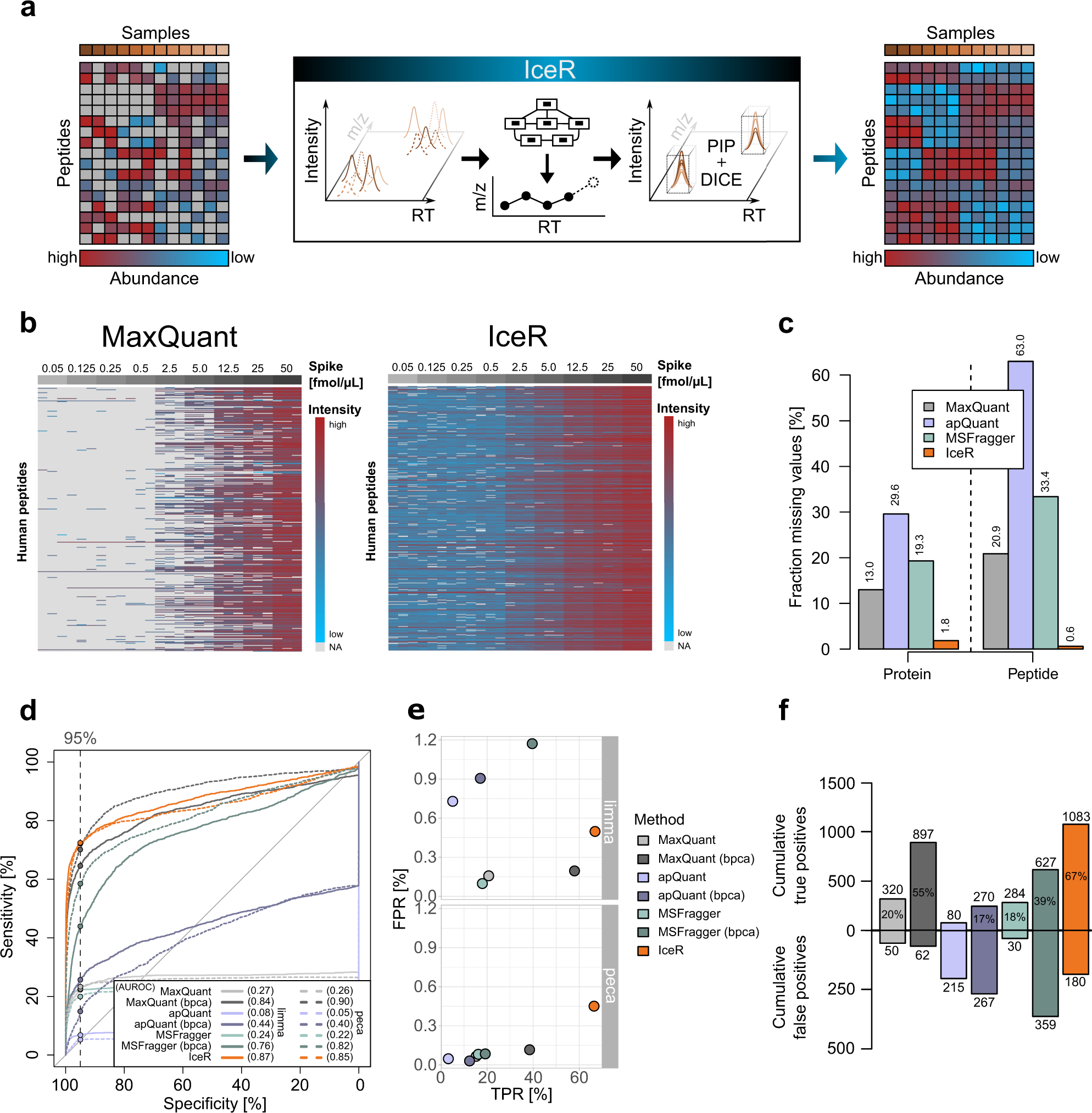
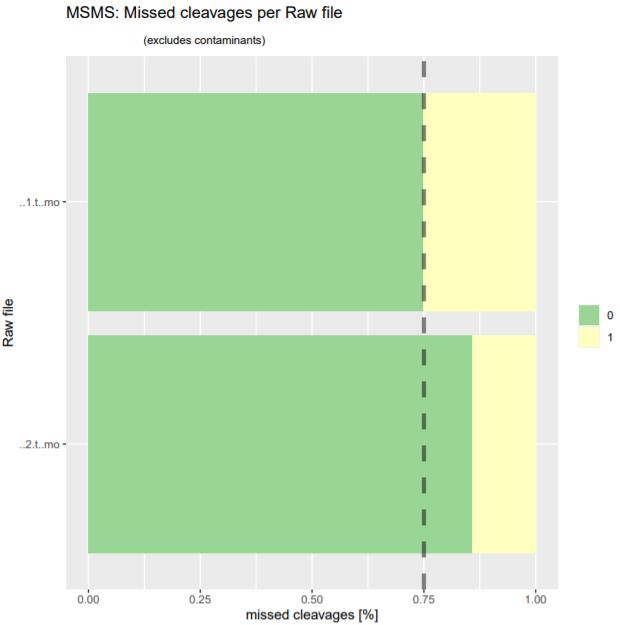









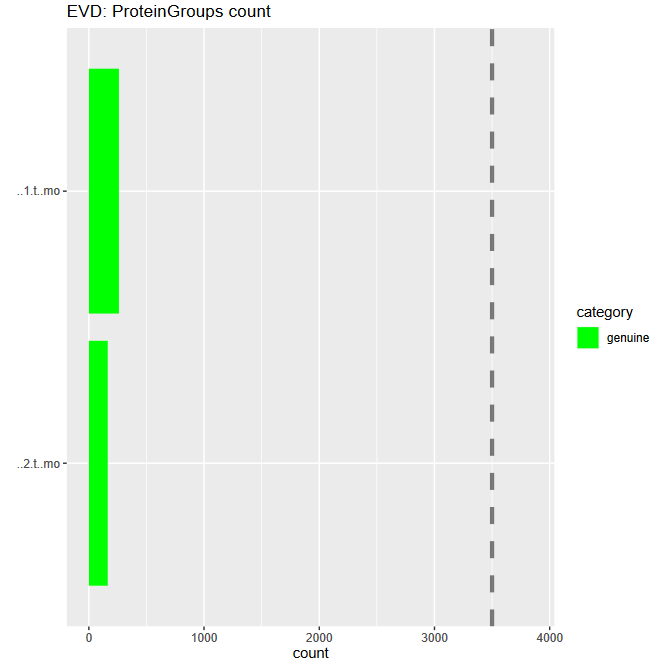




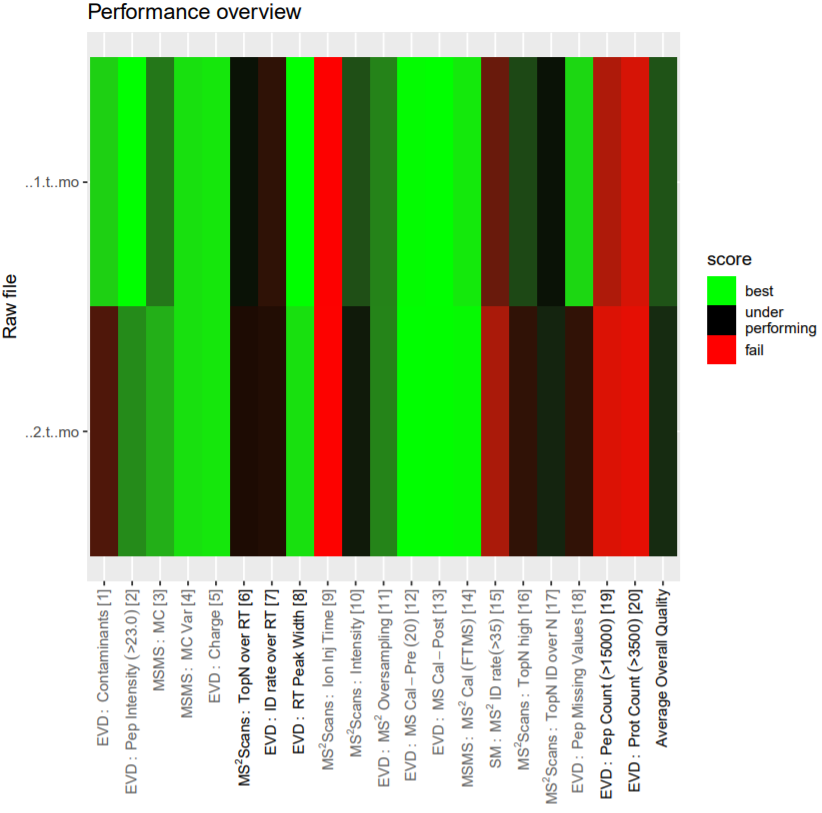

Post a Comment for "41 maxquant label free quantification"